-Bacstain- Series

-Bacstain- Series
Microbial staining reagent(-Bacstain- Series)
Colony formation using an agar plate is a very common and reliable method for counting bacterial cells. However, it takes quite a long time to form a colony. Therefore, alternative detection methods have been developed. Bacteria-specific gene amplification methods such as PCR, LAMP, and nucleus staining are quite rapid, but these methods count dead bacteria as well. Therefore, detection of live cell functions is essential to determining the actual number of living bacteria in a sample. Tetrazolium salts can be used to detect respiratory activity of bacterial cells or mitochondria. CTC is a tetrazolium salt and is reduced by this respiratory activity to form fluorescent CTC formazan on the cell surface. Therefore, CTC is used for specific staining of aerobic live bacteria and can be applied to hard-to-culture bacteria (VNC: viable but non-culturable). CTC forms a fluorescent formazan by an electron transfer system. However, CTC alone is not sensitive enough to stain single cells. Therefore, the CTC-Rapid Staining Kit contains an enhancing reagent that improves the CTC staining efficiency. Compared with staining with CTC only, this staining kit enables rapid and sensitive staining of microorganisms. Maximum wavelengths of the CTC formazan dye are 430 nm or 480 nm for excitation and 630 nm for emission.
Microbial staining reagent(-Bacstain- Series)
10 μl, 1000 μl pipettes, incubator, Microscope (blue excitation filter and red emission filter) or flow cytometer (488 nm blue laser)
Microbial stain reagent (-Bacstain- series)
Bacterial and fungal viability is generally assessed by colony forming ability on agar medium, but this procedure requires a long time (24-72 hours). In addition, it is said that most of the microorganisms existing in the environment have not yet found the optimum culture conditions. Rapid detection methods are also evolving, and genetic tests such as PCR method and LAMP method are also widely used. However, these genetic tests also detect dead cells, so survival cannot be determined.
Fluorescent staining, like the -Bactain- series, is also applied to survival measurement as one of the rapid detection methods.
| Product code | Product name | Package | SDS / Protocol |
|---|---|---|---|
| BS01 | -Bacstain- CTC Raoud Staining Kit (for Flow cytomery) | 100 assays | SDSProtocol |
| BS02 | -Bacstain- CTC Raoud Staining Kit (for Microscopy) | 100 assays | SDSProtocol |
| BS03 | -Bacstain- CFDA solution | 100 assays | SDSProtocol |
| BS04 | -Bacstain- DAPI solution | 100 assays | SDSProtocol |
| BS05 | -Bacstain- AO solution | 100 assays | SDSProtocol |
| BS07 | -Bacstain- PI solution | 100 assays | SDSProtocol |
Product information
Microbial staining reagent(-Bacstain- Series)
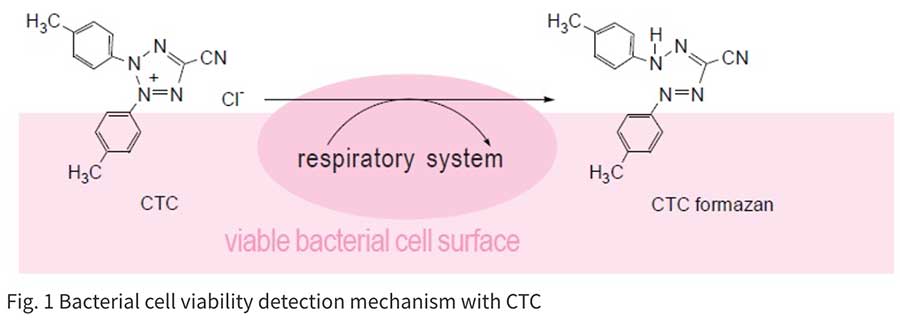
Technical info
Microscopy detection
1. Centrifuge bacteria culture and remove the supernatant, and then resuspend the bacteria pellet with PBS(-).
2. Add CTC + Enhancing reagent-B. Incubate at 37°C for 1 hour.
3. Prepare a slide and detect fluorescence by B-excitation filter set.
Flow cytometry detection
1. Centrifuge bacteria culture and remove the supernatant, and then resuspend the bacteria pellet with PBS(-).
2. Add CTC + Enhancing reagent-A. Incubate at 37°C for 1 hour.
3. Analyze the cells with a flow cytometry: 488 nm excitation, 630 nm emission.
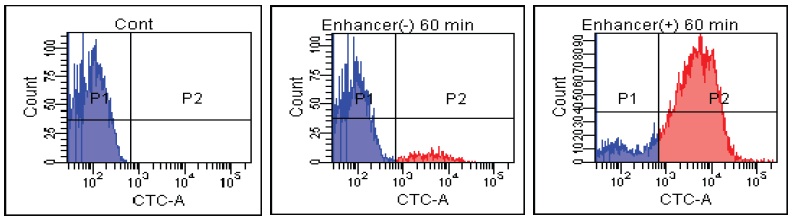
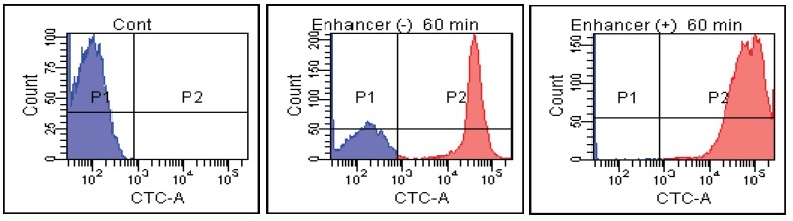
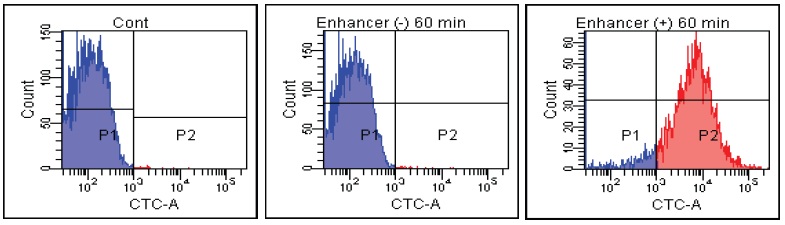
FACS Data
E. coli
B. Subtilis
Candida albicans
E. faecalis
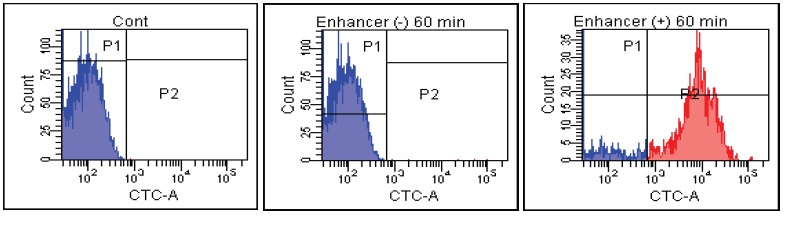
Fig. 2 CTC staining of various microorganisms with and without Enhancing reagent.
Experimental Example
Observation by Microscopy
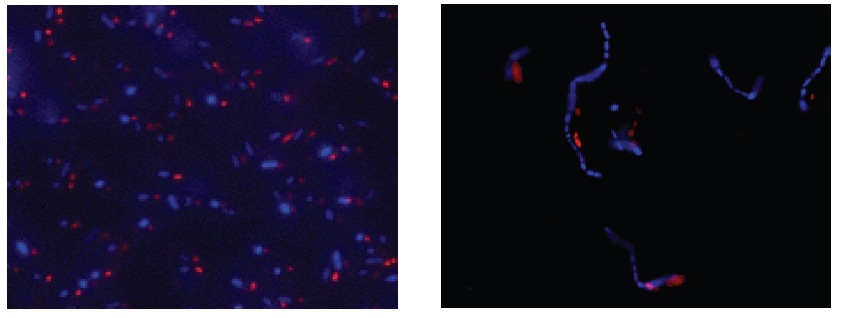
Fig. 3 E. coli staining (left) and L. casei staining (right) with CTC and DAPI. Bacterial cells were stained with CTC Rapid Staining Kit first, and then 1 μl of DAPI solution was added. The cells were incubated at room temperature for 5 min. Formaldehyde fixation with 1-4% formaldehyde can be performed before DAPI staining.
References
2) E. Severin, J. Stellmach and H. -M. Nachtigal, "Fluorimetric Assay of Redox Activity in Cells", Anal. Chim. Acta, 1985, 170, 341.
3) G. G. Rodriguez, D. Phipps, K. Ishiguro and H. F. Ridgway, "Use of a Fluorescent Redox Probe for Direct Visualization of Actively Respiring Bacteria", Appl. Environ. Microbiol., 1992, 58(6), 1801.
4) G. Schaule, H. C. Flemming and H. F. Ridgway, "Use of 5-Cyano-2,3-ditolyl Tetrazolium Chloride for Quantifying Planktonic and Sessile Respiring Bacteria in Drinking Water", Appl. Environ. Microbiol., 1993, 59(11), 3850.
5) R. A. Bovill, J. A. Shalloross and B. M. Markey, "Comparison of the Fluorescent Redox Dye 5-Cyano-2,3-ditolyltetrazolium Chloride with p-Iodonitrotetrazolium Violet to Detect Metabolic Activity in Heat-stressed Listeria monocytogenes Cells", J. Appl. Bacteriol., 1994, 77(4), 353.
6) M. T. E. Suller and D. Lloyd, "Flow Cytometric Assesment of the Postantibiotic Effect of Methicillin on Staphylococcus aureus", Antimicrob. Agents Chemother., 1998, 42(5), 1195.
7) M. Kawai, N. Yamaguchi and M. Nasu "Rapid Enumeration of Physiologically Active Bacteria in Purified Water Used in the Pharmaceutical Manufacuturing Process", J. Appl. Microbiol., 1999, 86, 496.
8) N. Yamaguchi, M. Sasada, M. Yamanaka and M. Nasu, "Rapid Detection of Respiring Escherichia coli O157:H7 in Apple Juice, Milk and Ground Beef by Flow Cytometry", Cytometry , 2003, 54A, 27.
9) A. Kitaguchi, N. Yamagauchi and M. Nasu, "Enumeration of Respiring Pseudomonas spp. in Milk within 6 Hours by Fluorescence In Situ Hybridization Following Formazan Reduction", Appl. Environ. Microbiol., 2005, 71(5), 2748.
10)A. Hiraishi and N. Yoshida, "An Improved Redox Dye-Staining Method Using 5-Cyano-2,3-Ditoryl Tetrazolium Chloride for Detection of Metabolically Active Bacteria in Activated Sludge", Microbes Environ., 2004, 19(1), 61.