XANTHINE DEHYDROGENASE [XDHⅡ]
from Microorganism
(Xanthine: NAD+ oxidoreductase, EC 1.17.1.4)
Xanthine + NAD+ + H2O → Urate + NADH + H+
Preparation and Specification
- Appearance
- : Brownish solution
- Specific activity
- : More than 100 U/ml
Properties
- Substrate specificity
- : See Table 1
- Molecular weight
- : 240 kDa (TSK G–3000SW gel filtration)
- Isoelectric point
- : pH 4.5±0.2
- Michaelis constants
- : Xanthine 2.4 × 10–4M
NAD+ 7.5 × 10–5M
Thio–NAD+ 8.0 × 10–5M
- Optimum pH
- : 8.5Figure 1
- pH stability
- : 6.5–9.5 (45℃, 15 min) Figure 2
- Optimum temperature
- : 55℃ (Tris–HCl buffer) Figure 3
- Thermal stability
- : Stable at 60℃ and below (pH 7.5, 15 min) Figure 4
- Effect of various chemicals
- : See Table 2 and Table 3
Applications for Diagnostic Test
This enzyme is useful for enzymatic determination of inorganic phosphate when coupled with purinenucleoside phosphorylase (T–69)
| PNPL Ⅱ | ||
| Pi + Inosine | → | Hypoxanthine + Ribose 1-phosphate |
| XDH Ⅱ | ||
| Hypoxanthine + 2 NAD+ + H2O | → | Urate + 2 NADH + 2 H+ |
Table 1. Substrate specificity
| Substrate (1mM) | Relative activity (%) |
|---|---|
| Hypoxanthine | 100 |
| Xanthine | 75 |
| Uric acid | 0 |
| Adenosine | 0 |
| Inosine | 0 |
| Xanthosine | 0 |
| Guanosine | 0 |
| Adenine | 0 |
| Guanine | 0 |
| 8–Azaxanthine | 0 |
| 8–Azaguanine | 0 |
Table 2. Effect of metal ions on XDH Ⅱ activity
| Metal ion | Relative activity (%) |
|---|---|
| None | 100 |
| NaCl (10mM) | 100 |
| KCl (10mM) | 100 |
| NH4Cl (10mM) | 100 |
| CsCl (10mM) | 100 |
| LiCl (10mM) | 100 |
| CaCl2 (1mM) | 100 |
| BaCl2 (1mM) | 92 |
| MgCl2 (1mM) | 100 |
| MnCl2 (1mM) | 88 |
| NiCl2 (1mM) | 58 |
| CuCl2 (1mM) | 50 |
| CoCl2 (1mM) | 92 |
Table 3. Effect of detergents on XDH Ⅱ activity
| Detergent (0.5%) | Relative activity (%) |
|---|---|
| None | 100 |
| Pluronic L–71 | 100 |
| Adekatol NP–690 | 100 |
| Adekatol PC–8 | 100 |
| Nikkol NP–18TX | 100 |
| Tween 80 | 100 |
| Triton X–100 | 100 |
Fig.1 pH Optimum
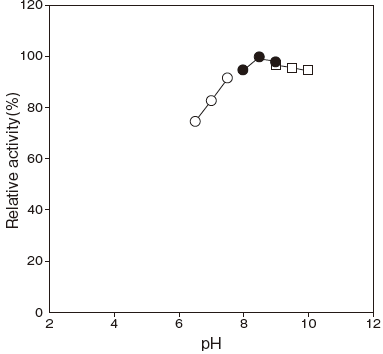
●: Tris-HCI buffer
□: Glycine-NaOH buffer
Fig.2 pH Stability
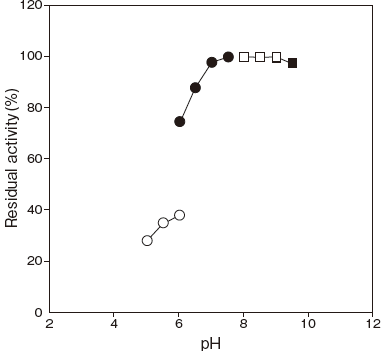
〇: Acetate buffer
●: Phosphate buffer
□: Tris-HCI buffer
■: Glycine-NaOH buffer
Fig.3 Optimum Temperature
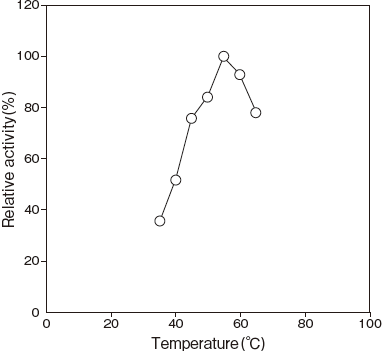
100 mM Tris-HCI buffer
Fig.4 Thermal Stability
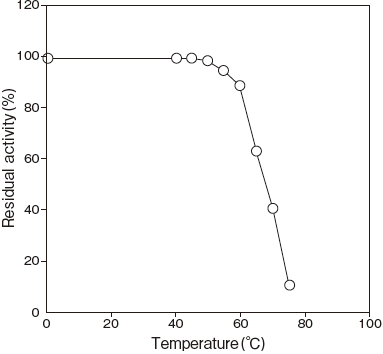
100 mM Tris-HCI buffer
Assay
Principle
The assay is based on the increase in absorbance at 340 nm as the formation of NADH proceeds in the following reaction:
| XDH Ⅱ | ||
| Xanthine+NAD++H2O | → | Uric acide+NADH+H+ |
NAD : Nicotineamido adenine dinucleotide
Unit definition
One unit is defined as the amount of enzyme which converts 1 μmole of xanthine to uric acid per minute at 37℃ under the conditions specified in the assay procedure.
Reagents
- Reaction mixture
1M Tris–HCl buffer pH9.0 0.30ml 10mM NAD solution 0.60ml 10mM Xanthine solution pH 10.5 ± 0.5 1) 0.60ml
Distilled water 1.50 ml 1) : 10mM Xanthine solution pH 10.5 ± 0.5
Dissolve 152 mg of xanthine with 80 ml of distilled water, adjust pH to 10.5 ± 0.5 at 25℃ with 1 N NaOH and add distilled water to make a total of 100 ml.
- Enzyme dilution buffer
1M Tris–HCl buffer pH 8.0 - Reagents
NAD: NACALAI TESQUE, INC. #24334–84
Xanthine: FUJIFILM Wako Pure Chemical Corporation#241–00013Tris (hydroxymethyl) aminomethane:Sigma Chemical Co. #T–1503
Enzyme solution
Dilute accurately 0.5 ml of the sample with enzyme dilution buffer to make a 50–fold solution. Dilute it with enzyme dilution buffer to adjust the concentration as required.
Procedure
- Pipette accurately 3.0 ml of reaction mixture into a small test tube and preincubate at 37℃.
- After 5 min, add 50 μl of enzyme solution and mix to start the reaction at 37℃.
※ In the case of a test blank, add 50 μl of enzyme dilution buffer in place of enzyme solution. - After starting the reaction, measure the rate of increase per minute in absorbance at 340 nm. The rate must be measured within the linear portion of the absorbance curve.
△ A/min = (As/min−Ab/min) ≦ 0.070 Abs/minAbsorbance sample : As/min blank : Ab/min
Calculation
- Activity (U/ml) = {(△A/min)/(6.22)} × 3.05/0.05 × D
6.22 : millimolar extinction coefficient of NADH at 340 nm (cm2/ μmole)3.05 : final volume (ml) 0.05 : volume of enzyme solution (ml) D : times of dilution in enzyme solution
Storage
Storage at −20℃ in the presence of a desiccant is recommended.
XDH Ⅱ活性測定法 (Japanese)
試薬液
- 反応試薬混合液
1M トリス−HCl 緩衝液 pH9.0 0.30ml 10mM NAD 溶液 0.60ml 10mM キサンチン溶液 pH10.5 ± 0.5 1) 0.60ml 精製水 1.50ml 1) : 10mM キサンチン溶液 pH10.5 ± 0.5
キサンチン152mg を精製水80ml で溶解した後1N NaOH でpH10.5 ± 0.5 (25℃) に調整し、精製水で全容100ml とする。 - 酵素溶解希釈用液
1M トリス−HCl 緩衝液 pH8.0 - 試薬
NAD (ニコチンアミドアデニンジヌクレオチド) :ナカライテスク製 #24334–84キサンチン (C5H4N4O2) :富士フイルム和光純薬製#241–00013トリス (ヒドロキシメチル) アミノメタン:シグマ製 # T–1503
酵素試料液
- 検品0.5ml を酵素溶解希釈用液で50 倍に希釈する。
その液を酵素溶解希釈用液で適宜希釈する。
測定操作法
- 小試験管に反応試薬混合液3.0ml を正確に分注して37℃で予備加温する。
- 5 分経過後、酵素試料液50 μl を加えて混和し、37℃で反応を開始する。
※ 盲検は酵素試料液の代わりに酵素溶解希釈用液50μl を加える。 - 反応開始後、340nm における吸光度を測定して直線的に反応している1 分間当たりの吸光度変化を求める。
求められた吸光度変化を試料液はAs/min、盲検液はAb/min とする。
ΔA/min = (As/min−Ab/min) ≦ 0.070 Abs/min
計算
活性 (U/ml) = {(ΔA/min)/(6.22)} × 3.05/0.05 × D| 6.22 : | NADH の340nm におけるミリモル分子吸光係数 (cm2 / μmol) |
| 3.05 : | 反応総液量 (ml) |
| 0.05 : | 反応に供した酵素試料液量 (ml) |
| D : | 酵素試料液の希釈倍率 |
